Structure-Based Modeling of Photosynthetic Light Harvesting
Understanding light harvesting in photosynthetic proteins through physics-based Hamiltonian construction rather than empirical parameter fitting
The Challenge
Random Assignment Approach
Traditional methods assign random site energies to pigments and attempt to fit experimental spectra through parameter adjustment. While this approach might produce matching spectra from millions of combinations, it doesn't provide genuine understanding of the underlying photosynthetic system.
"Matching experimental data ≠ Understanding the system"
Our Structure-Based Solution
We develop structure-based Hamiltonians where site energies are calculated from interactions with the surrounding protein environment. This approach ensures that environmental changes directly affect pigment energies, providing mechanistic insight into photosynthetic function.
"Structure-based modeling reveals the 'why' behind the spectra"
MCCE/CDC Pipeline
Our computational pipeline combines Multi-Conformer Continuum Electrostatics (MCCE) with Charge Density Coupling (CDC) methods to construct physically meaningful Hamiltonians for photosynthetic complexes.
Protein Structure Analysis
Starting with high-resolution crystal structures, we identify all ionizable residues and their potential conformations within the protein environment.
Conformer Generation (MCCE)
Generate all possible side-chain conformations and protonation states for ionizable residues. Calculate pKa values based on electrostatic interactions with the surrounding environment.
Monte Carlo Sampling
Use statistical sampling to determine the most energetically favorable conformations, creating the 'most occupied structure' that represents the equilibrium state.
Site Energy Calculation (CDC)
Calculate the site energy of each pigment based on electrostatic interactions with the optimized protein environment, accounting for local electric fields and polarization effects.
Spectral Prediction
Generate absorption and fluorescence spectra for the protein complex, enabling direct comparison with experimental measurements and validation of the Hamiltonian.
Key Achievements
CP26 & CP24 Hamiltonians
First-ever construction of structure-based Hamiltonians for CP26 and CP24 minor antenna complexes of Photosystem II
Enhanced Spectral Accuracy
Achieved quantum mechanics-comparable accuracy in predicting absorption and fluorescence spectra with significantly reduced computational cost
Protonation State Impact
Demonstrated critical role of non-standard amino acid protonation states in hydrophobic protein cores affecting pigment energetics
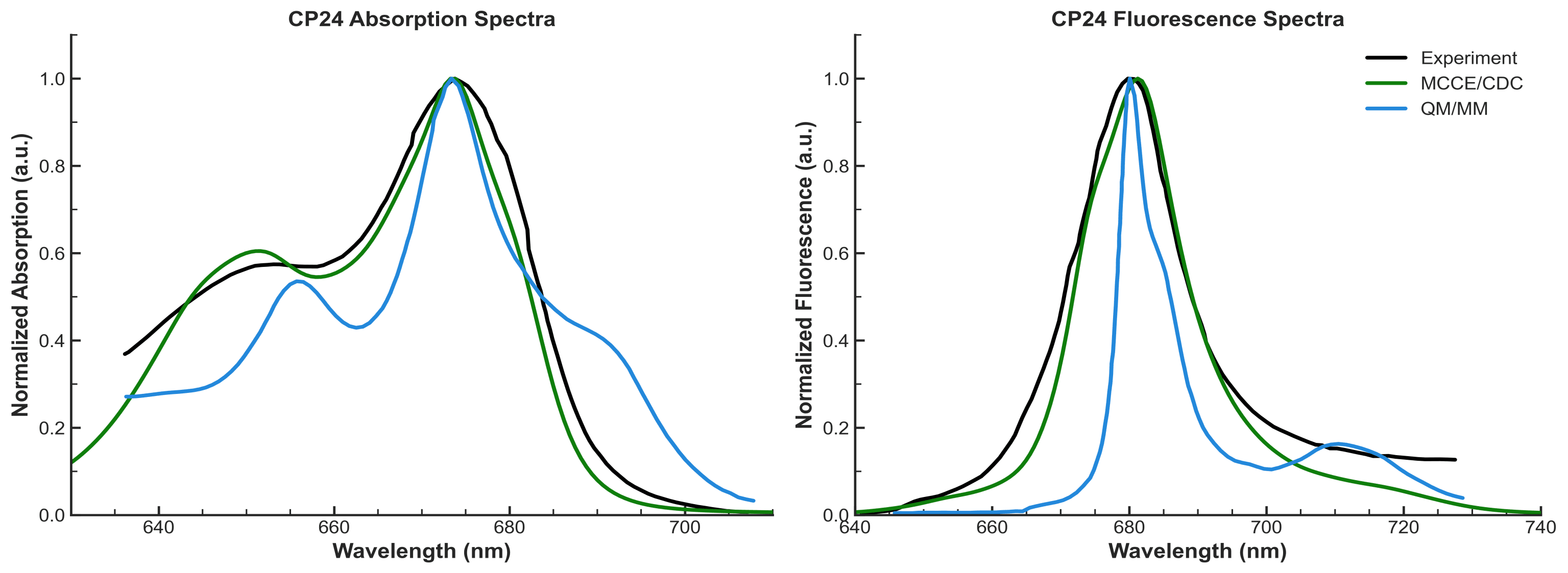
Comparison of experimental spectrum (black) with simulated spectra of the CP24 minor antenna complex. The MCCE/CDC-based simulation (green) is generated using our structure-derived Hamiltonian pipeline, while the QM/MM (blue) represents a conventional QM/MM approach. Our method accurately reproduces key spectral features, including peak position and broadening, demonstrating its effectiveness in modeling pigment-protein excitonic interactions.
Computational Approaches Comparison
Two distinct frameworks for modeling protein electrostatics and dynamics
MCCE/CDC Framework
Focus: Charge State Optimization
Models discrete side-chain positions and their charge (protonation) states. The protein backbone is typically held rigid while optimizing electrostatic interactions.
Method: Monte Carlo Sampling
Uses statistical sampling to find the most energetically favorable conformations at equilibrium, determining optimal protonation patterns.
Output: Equilibrium Properties
Calculates thermodynamic properties like pKₐ values at specific pH conditions. Answers "what is the most stable state?" rather than dynamic processes.
Molecular Dynamics/CDC Framework
Focus: Molecular Motion
Models the explicit movement of every atom in the entire system, including protein, water, and ions over time.
Method: Equations of Motion
Simulates continuous motion by calculating forces on all atoms at femtosecond time steps, following Newtonian mechanics.
Output: Dynamic Trajectory
Shows how the system evolves over time, revealing conformational changes, binding events, and kinetic processes.
Computational Cost Comparison
Typical computational time required for modeling a single photosynthetic protein complex
MCCE/CDC Framework
Runs efficiently on local workstations or small clusters
MD/CDC Framework
Requires high-performance computing clusters with hundreds of cores
Scale represents relative computational cost - MCCE enables rapid Hamiltonian construction compared to MD simulations
Deep Dive: Interactive Spectra Theory
Explore the theoretical foundations behind our structure-based modeling approach. This interactive guide breaks down the quantum mechanical principles, mathematical formulations, and computational methods used to construct photosynthetic Hamiltonians.
🔬 Interactive Theory Explorer
Navigate through 5 comprehensive sections covering Frenkel exciton theory, Hamiltonian construction, and disorder modeling
📊 Live Data Visualization
Interactive charts and heatmaps showing site energy calculations and excitonic coupling matrices from our CP29 research
⚛️ Mathematical Framework
Detailed equations and explanations of each Hamiltonian component with interactive hover explanations
Interactive tutorial based on Müh et al. (2014)
Goal: Visualizing Energy Transport
The culmination of this research is a detailed, structure-based understanding of how excitation energy flows through the Photosystem II complex. By modeling interactions and calculating transport rates, we can visualize the precise pathways energy takes between protein subunits. This provides critical insights into the efficiency and regulation of light harvesting.

Atomistic model illustrating energy pathways in a light-harvesting complex (PSII). Each color represents protein chains and arrows show the rate of excitation energy transport between complexes in picoseconds.